Let’s face it. Exons get all the glory. They’re the star players of the game, the quarterbacks, the offensive line. Meanwhile, introns and other intergenic regions play a more defensive strategy, are vitally important, yet rarely get their faces on sports cards, so to speak.
This whole notion of “junk DNA” still sours the scientist’s imagination even though it’s dawning on us that there is an elegant beauty and usefulness to these once-ignored segments. As an example of their importance from my own field of autism research, when you take a look at the variety of mutations which find association with the condition– even the big ones like CNTNAP2— very rarely do these mutations occur in exons but instead occur within intronic regions. We can’t yet say whether all these different mutations lead to alterations in gene expression or whether there is something more complex going on, but it certainly begs the question and suggests a more important role for introns than was once believed.
For those who follow this blog, you may be aware that I’m fascinated by transposable elements (TE). I can’t quite pinpoint what I find so fascinating except that 1) there’s so many TEs in the genome that it’s absolutely astounding, and 2) they largely seem to be ignored in the field of genetics– which I find rather baffling. At most they tend to be treated as an interesting phenomenon but are not foundational to broader genetics theory– even though the entire development of transgenic animal lines for research hinges on transposition. While I don’t expect every geneticist to dedicate his or her academic life to focusing on mobile elements, the fact that so many seem so uninterested is pretty sad. It’s tantamount to working in the field blinded to an entire range of effects. Just think how genetics theory may currently be closed off to alternative interpretations of data!
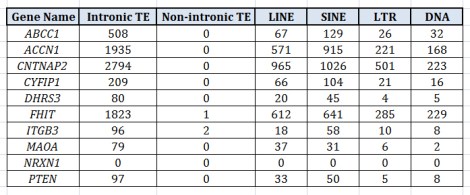
One thing which I find really interesting about TEs is that, by and large, they tend to be relegated to intronic regions. The table below is a few examples pulled from my own work, showing the four major categories of TEs in humans and the number of TEs in intronic regions of a given gene. A correlation between the categories, LINE and SINE, is to be somewhat expected since (autonomous) LINEs produce protein products which make it possible for (nonautonomous) SINEs to insert themselves into the DNA. But note that the LINEs, SINEs, LTRs, and DNA tranposons all correlate well with one another. To me it suggests there is something unique to these intronic regions that, when they attract one type of mobile element, they generally attract all of them.
At first glance, we may look at this tendency toward intronic insertion as something “adaptive”, i.e., if TEs targeted exonic, promoter, or enhancer regions with greater frequency, that would be maladaptive and those cells would more likely die off; therefore, those cells which allowed the insertion of TE content into the intronic regions, while possibly altering gene expression in some ways, were not as detrimentally affected and therefore lived to pass on this tendency to their progeny.
On some level, this may well be true. But for my part, I also wonder whether there isn’t something particular about the nature of intronic sequences which attracts transposition in the first place.
In previous blogs, I have stated that contrary to how most of us have been taught to imagine the genome (an analogue sequence), its 3D formation is vital to how the genome works. –In fact, I shouldn’t even say “vital” in the sense it is one part of a larger whole. DNA works the way it does, just as proteins do, in large part because of its 3D conformation. And as the term “conformation” implies, it is a dynamic form. It’s doesn’t remain in a complacent double-helical state but continually changes its superhelical twist, and parts of it jut out into alternative conformations like triple helices, cruciforms, and hairpins, in reaction to tension, binding partners, and the like. While the literature tentatively suggests it, I have suspicions that some of these hairpins and cruciforms may actually provide targets for transposons. Call it a hunch. Because such events, in relation to transposition and even daily events like transcription, are described in the literature over and over again. Too often to be coincidence. There’s something useful about these alternate forms and some day I’m sure we’ll know what it is.
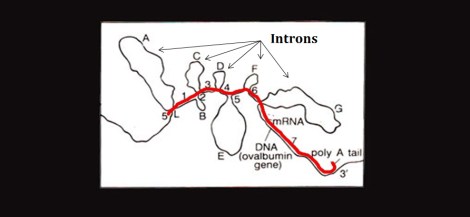
Given my interest in alternative conformations, I came across an image in the Color Atlas of Genetics, 4th Ed. which struck me as fascinating. I think I had seen something like it way back when but until now it hadn’t really caught my attention. It was an electron micrograph of the hybridization of the original Ovalbumin DNA sequence and one of its RNA transcripts. From the image, you can see that the transcript has only hybridized with the exons of the DNA while the introns jut out and form a loop formation. The red line indicates the RNA being transcribed:
While the entire sequence is originally transcribed, the introns are spliced out post-transcriptionally. But there is growing evidence to suggest that intronic DNA segments, especially those containing palindromic and other repetitive sequences, are capable and perhaps prone to hairpin formation [1]. The loop formation above is not the “hairpin” formation I’m referring to; nevertheless, this figure got me thinking and putting words to the ideas that’ve been swimming around my head for the last half year.
Loops. Atypical conformations. Maybe these non-double helical events, these hairpins, these random coils, are all targets for transposable elements. For what reasons I don’t know. Perhaps in these conformations, the base-pairing bonds are fewer and weaker than those in the double helix. Because of this, maybe they’re less stable and more easily broken. Thus, perhaps they are simply “weak spots” in the gene, more prone to insertions, point mutations, and copy number variants. –Is it also not a surprise then that both V(D)J recombination and certain transposition events actually utilize a hairpin structure themselves to create double-stranded breaks? [2]
It’s just a hunch. There’s a lot of tantalizing evidence out there right now, with no doubt more to come, and I genuinely look forward to its synthesis. Time will tell what functions these alternate conformations play. In any case, it will lend towards a more complex and 3-dimensional understanding of how the genome works and, when mutations do occur, why that is.
As an aside, I hope some day this concept of “random” mutation is disposed of. Who ever heard of anything truly random in nature anyways? Isn’t the very premise of science based on the assumption that everything within the natural world has an explanation and is therefore predictable? “Random mutation”, ha! Silly, Rabbit, randomness doesn’t belong in science. 🙄

Great piece, Emily. In some ways I reckon the HGS is a real obstacle to understanding – because it projects a 2D string of operational units, which is so the wrong image to retain in ones thought. Obviously the chromosomes are 3D and, in reality 4D because they’re not fixed in time or operational status.
Perhaps rather than “random” there are inevitable “stress points” which in time give way, so allowing mutational interpolations. We observers cannot, of course, predict these stress points so they appear random – at best we can estimate how frequently they will occur.
Thanks, greencentre. 🙂 And I agree re the randomness thing. Some of the work I’m currently leaning into involves looking at “weaker genes” in relation to their mutational tendencies. But I totally agree about having a 2D rendering of DNA as “the wrong image to retain in one’s thoughts.” How we cognitively model our theories are fundamental to how we interpret our data.
Here I’m going to shamelessly steal an idea, an insight, that springs directly from?–you guessed it, yes–the work of Pierre Sonigo and Jean-Jacques Kupiec. I hope that your better knowledge of these matters will, if possible, discern whatever pertinence there may be what follows –though I may be mistaken about how pertinent this comment is, your post here recalls these thoughts for me:
first, a citation from Kupiec and Sonigo, “Ni Dieu, ni gène” (Neither God nor gene”) at page 214:
“Pour vivre, les animaux ont besoin de nouriture plus que d’information.”
That is, “In order to live, animals (this, your translator’s note: and, we might add, living cells, no less so) need food more than they need information.”
There could be some value in applying at least the lowest levels of Abraham Maslow’s hierarchy of needs to the cell in an examination of its life processes. So, while cells probably don’t rise to the level of needing “Love/belonging” or “esteem” or “self-actualization”, they do “need” most of the lowest level’s elements: breathing, food, water, homeostasis and excretion.
To that prologue, I want to tie this, one of the most fundamental of ideas in the work of Kupiec– namely, the alternatives of “stable”/”stabilization” on one hand, and, “instable”/”instability” on the other. From reading their work, it is easy for the naive reader to draw an inference, whether they intend it or not, though I believe that they definitely do intend the reader grasp this idea:
Both homeostasis or stability and its opposite, “Enantiostasis” (see C. P. Mangum & D. W. Towle (1977). “Physiological adaptation to unstable environments”) can be seen as arising naturally from the given circumstances of the cell’s immediate environment or, in the case of more complex organisms, the plant or animal environment.–those circumstances themeselves being heavily influenced by chance–though we know of course that such a thing as randomness never occurs in nature, for just a moment, we’re going to prentend that it might. ;^)
So, what might occur to the naive reader is the intriguing idea that mutation and all processes associated with it, including the occurrance of these fascinating loops and other physical structures, are in fact the direct results of cells (or plant and animal life) in the condition of enantiostasis. That is, simply, enantiostasis automatically provokes or prompts mutation as a physical stimulus-response reaction and, moreover, does this ‘blindly’, without the cell’s knowledge or awareness as a prerequisite. That may be due to the fact, cited above, that “In order to live, animals need food more than they need information.”
Again, the naive reader could find himself or herself drawn to speculate: perhaps “mutation” is simply the reactive outcome of enantiostasis at any and all levels of complexity. The particular sources of enantiostasis are without number. They can be virtually anything in the environment that upsets homeostasis, or the regular and routine capacity of the cell or organism to acquire its material requirements for existence–food, water, respiration, excretion, etc. A prairie fire can induce enantiostasis; a drought, a flood, or, in the body’s interior environment, anything that intervenes to produce a divergent shortage or oversupply of these material necessities of life for the cell may be interpreted to produce a a physicalreaction in which we see what is called “mutation” in process. A new instance of disruption in the local environment, and spontaneous disruption in the cells’ otherwise routine processes.
Then, going further, suppose that the disruptions in the cell’s processes happen–again, not by design but by sheer random happenstance–to produce a revision of the cell’s (or cells’) physical presentation, and by chance, one or more of these alterations produces a beneficial effect which restores the cellular environment to a new-found homeostasis? That could be viewed in microcosim as a tiny insance of cellular natural selection at work in the living environment and it is this aspect, this manner of viewing the situation which Kupiec and Sonigo suggest as in part the keys to a new (or, to a return to Darwin’s original insights about nature and evolutionary processes) view of living processes at all levels.
I am one of these naive readers in whom Kupiec and Sonigo have inspired such a view.
For me, a few much-appreciated and striking elements from your post:
“Loops. Atypical conformations. Maybe these non-double helical events, these hairpins, these random coils, are all targets for transposable elements.”
“How we cognitively model our theories are fundamental to how we interpret our data.”
and, with irony aplenty, “As an aside, I hope some day this concept of “random” mutation is disposed of. Who ever heard of anything truly random in nature anyways? Isn’t the very premise of science based on the assumption that everything within the natural world has an explanation and is therefore predictable? “Random mutation”, ha! Silly, Rabbit, randomness doesn’t belong in science. 🙄 ”
So, here’s my offer for return irony, with thanks,
🙄
Postscript the above citation of Governor Berkeley is cited in Neil Postman & Charles Weingartner, Teaching as a Subversive Activity, 1969, Delacorte Press.
>>So, what might occur to the naive reader is the intriguing idea that mutation and all processes associated with it, including the occurrance of these fascinating loops and other physical structures, are in fact the direct results of cells (or plant and animal life) in the condition of enantiostasis.<<
I like the way you've framed these concepts. These loops are undoubtedly results of the continually changing conformation of the local DNA. For instance, it has been documented that loops, random coils, etc. often occur in higher frequency when the supercoil is coiled with greater tensional strain. It's as if one aspect of homeostasis to the B-helix is that a certain level of energy is used to maintain the homeostatic configuration (a concept not unrelated to entropy in physics). However, when a greater strain is placed upon the local DNA, in the form for instance of a transcriptional activator or some other regulatory element, then it becomes harder and harder (in line with the level of local tension) to keep these loops and coils from occurring more frequently.