You may have noticed that S.O.A.C. has been slow for the last few weeks. Well, I’m pleased to reported that my husband and I have just moved shop to Greenville, South Carolina, where we’ve started work at an extended campus of the University of South Carolina, in conjunction with the Greenville Health System. We’re both excited for the change and are super-anxious to get settled in and working on more research. Hopefully I’ll have some good work to report on in future!
Anyways, this week I want to report on a recent article that appeared in Nature Genetics by Krumm et al. (2015), addressing trends in rare copy number and single nucleotide variants. While I won’t completely summarize the work, the team did report a few interesting findings.
Krumm et al. studied genetic trends from the Simons Simplex Collection (SSC), which includes only families with single cases of autism. Though they found no significant differences in the transmission of likely gene-disrupting (LGD) and missense mutations when comparing those with autism and their unaffected siblings, when they looked at genes that tend to be more intolerant to mutation events, there were significant differences between autistics and their siblings and between affected and unaffected families. In short, though the overall rate of LGDs didn’t vary between autistics, siblings, and controls, when looking at certain genes that show stronger conservation rates throughout the human population, LGDs in these genes definitely showed links with simplex autism.
Interestingly, the team also looked at mutational burden related to diagnosis. Those with either a diagnosis of “autism” or “pervasive developmental disorder” were more likely to carry LGDs than those with a diagnosis of Asperger’s Syndrome, suggesting that LGD mutations may have strong links with autism severity. In support of this, those autistic individuals with an IQ between 70-100 showed a statistically significant genetic burden, whereas those with IQ 100+ did not.
Though these were all cases of simplex autism, there were families in which siblings rated similarly to their autistic brothers or sisters on the social responsiveness scale (SRS) even though they did not have a diagnosis of autism. When Krumm et al. stratified their analysis in order to compare pairs of siblings with similar SRS scores with those who were more discordant, they found that autistic children from the discordant pairs had higher rates of LGDs. This agrees well with previous work that suggests that simplex cases of autism may exhibit higher rates of gene-disrupting mutations and may be more severe symptomologically compared to multiplex cases.
The team went on to study copy number variant (CNV) deletions and duplications, finding that, as previously reported, autistic children showed higher rates of CNVs compared to their unaffected siblings, which was driven primarily by deletion events. De novo (new) CNVs tended to be larger in autistics than in siblings and more often included genes that were less tolerant to mutations (i.e., more deleterious). Autistic children also showed enrichment for inherited CNVs compared to their siblings. These CNVs were more often inherited from the mother and tended to be smaller (less than 100 kilobases), suggesting a role for a Female Protective Effect in the mothers. In support of this notion, they also found that when studying single nucleotide variant (SNV) LGDs, male autistics were more likely to have inherited potentially deleterious mutations from the mother, whereas female autistics had a stronger enrichment in de novo mutations, once again suggesting that females on average require a heavier genetic burden in order to compare symptomologically to their male counterparts.
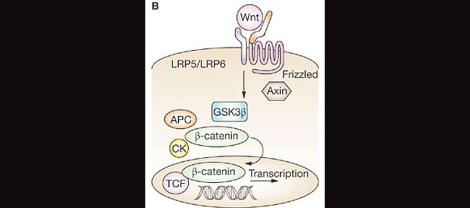
Associated with the de novo CNV events, Krumm et al. also found that the autism-risk genes, FMR1, associated with Fragile X Syndrome, and CHD8, associated with a currently unnamed autism-related syndrome, were overrepresented in these mutation events. Interestingly, small maternally-inherited duplication events were enriched as CHD8 target genes, though the same was not true for the fragile X protein, FMRP. They went on to study macrocephaly in those families with gene mutations potentially downstream of CHD8 activity, finding that both macrocephaly and microcephaly were overrepresented in this subset.
Individuals with CHD8 mutations. So far, almost all cases of CHD8 mutation studied have autism, indicating that CHD8 mutations are highly penetrant for the condition. A handful of other genes, including ADNP, show similar trends in penetrance.
This finding may agree well with recent work by Hormozdiari et al. (2015) that identified two large, non-overlapping functional networks linking autism candidate genes. One network focused around long-term potentiation and calcium signaling pathways, meanwhile the other is involved in histone modification, transcriptional regulation, and cell growth and differentiation. CHD8 is a primary player in the latter module, integral especially in regulating activity of the Wnt pathway.
While this newest study doesn’t provide any particularly miraculous leaps and bounds to the state of knowledge of autism genetics, it does offer a few new interesting tidbits as well as continue to reinforce blossoming concepts in the current literature, primarily involving the nature of genetic risk of autism, the differences in those risk factors between simplex and multiplex families and between high-, moderate-, and lower-functioning individuals, and between the sexes. In particular, once again, it supports the idea that rarer gene-disrupting mutations, be they CNVs or SNVs, may be more common to severe simplex cases of autism, meanwhile higher-functioning multiplex families could be characterized by more common variants that are comparatively less penetrant and confer lower, though still significant, risk for autism and may be more polygenic or even multifactorial, including environmental agencies. Time will tell.


Congratulations! Always exciting to start a new major chapter in life. Hope you get used to grits.
Thanks, Robert! I think I’ll be good with grits provided I can have ’em with cheese. 😉
The study was based on data from the Simons Simplex Collection SCC). Exclusionary criteria for acceptance in this study are: ‘Probands were excluded who were younger than 4 years of age or older than 18. Probands were also excluded for conditions that might compromise the validity of diagnostic instruments, such as nonverbal mental age below 18 months, severe neurological deficits, birth trauma, perinatal complications, or genetic evidence of fragile X or Down syndromes. My daughter would have been excluded for several reasons, exposure to a drug contraindicated for obstetrical delivery and neo complications (Asphyxia).
The SCC is meaningless to me and can only apply to the families studied and cannot be extrapolated beyond the group studied.
http://www.sciencedirect.com/science/article/pii/S0896627310008305
Thanks for the additional info. Yes, I can see how the SSC may not be well representative of all types of simplex autism. Good to keep in mind when interpreting (or using!) the data.
I would like to know the difference between CNVs and mutations. My impression is that mutations refer to an overall class of spontaneous (non-transmitted) changes in genetic material. CNVs, it would seem, are a type of mutation. Right or wrong?
That’s a little tricky to answer, Steven, in the sense that these terms are not always in full agreement with one another. The term “variant” shares links with the term “polymorphism”. In that instance, a polymorphism would be an allelic or copy number variant that occurs in > 1% of the population. If it occurs in 1% occurrence rate, just looking at rare CNVs. At that point, for sake of consistency, it would actually be nicer if they used the term “copy number mutation” or something similar, but hey, science is flawed by semantics. Hope that adds clarity for you in spite of the ambiguity.
CNV’s can be common and variably disributed throughout the general population. What do they do to ’cause’ autism. One possible explanation comes from AIDS researchers CNV’s in CC3L1 have been associated with HIV infection risk. A lower copy number is associated with an increased risk of HIV-1 infection, while a higher copy number is associated with reduced risk for acquiring HIV-1.
This would be consistent with ASD being a multifactorial condition and a gene x environment causality model
http://www.ncbi.nlm.nih.gov/pubmed/21209899
Robert, on that note, I think you’ll really like my next publication, though unfortunately I can’t talk too much about it here since it’s not published. But it deals with issues of penetrance and how less penetrant risk factors may prove to be much more multifactorial compared to, for instance, mutations in genes such as CHD8 and ADNP that have almost a 100% penetrance for autism. I’ll give you a heads up once it’s in press and will send it to you if you’re interested.
What also has to be kept in mind is that when a genetic mutation or variant is inherited the parent(s) are almost never affected certainly with respect to strict autism.
Though you may well see high-functioning or broader phenotype in the parents of some families.
Looking forward to your new study !!!
About penetrance. The study examine people with autism and claim 100% penetrance. CDH8 is also seen in patients diagnosed with cancer. The following study examined only cancer patients and report 100% penetrance for cancer. If you pooled both studies the 100% penetrance disappears for autism or cancer.
http://www.ncbi.nlm.nih.gov/pubmed/23835524
A final observation. The Krumm study did not have a control group from the general population. If it had the 100% penetrance would have also likely disappeared.
It’s my understanding though that every CHD8 mutation case they’ve been able to track down so far (which is currently 20+) fulfills full criteria for autism, which the exception of one case which was not available for testing.
Yes, but the cancer study reported on somatic mutations localized to the tumorous areas and the surround, which really doesn’t reflect on de novo germline mutations that affect the CNS as seen in CHD8-related cases.
All genetic mutations and common genetic variances are pleotropic with risk for multiple co-occuring medical conditions. Down syndrome has risk for intellectual disability, hearing loss, vision loss, heart defects, immune system problems, celiac disease as well as autism, all associated with the same chromosome mutation Trisomy 21. If you only recruit Down Syndrome patients with heart defects it is easy to claim Down syndrome has a 100% penetrance for heart defects. That’s the problem with the CDH8 study, they only included patients with autism and not people with the CDH8 variant who have gastric or small cell lung cancer, those without any medical problems but no autism.
http://ghr.nlm.nih.gov/condition/down-syndrome
No, I get what you mean, but I was under the impression that the cancer study involved somatic mutations that would not otherwise have an effect on brain development.