“An extremely large number of genes have been associated with autism. The functions of these genes span numerous domains and prove challenging in the search for commonalities underlying the conditions. In this study, we instead looked at characteristics of genes themselves, specifically in the nature of their transposable element content. Utilizing available sequence databases, we compared occurrence of transposons in autism-risk genes to randomized controls and found that transposable content was significantly greater in our autism group. These results suggest a relationship between transposable element content and autism-risk genes and have implications for the stability of those genomic regions” (Williams et al., 2013).
This month our publication, “Tranposable Elements Occur More Frequently in Autism-risk Genes: Implications for the Role of Genomic Stability in Autism,” was released in Translational Neuroscience. It’s the first in what we hope will be a line of investigations focusing on characterizing aspects of high-risk autism gene stability. To summarize, while most current research is focused on determining what these genes do in a variety of contexts and how they may ultimately lead to an autism phenotype, we instead are more interested in figuring out how and why these genes mutate in the first place.
Because our article may be a tad, shall we say, unapproachable for readers with little knowledge in this area of study, this blog post is primarily to serve as a “translation”. I hope this post can make things a little clearer and explain what this may all mean. But before I do any of that, I should first explain what a transposable element is for those who are unfamiliar.
Transposable elements (TE) are small sequences of the genome that are capable of some form of mobility. They are also often referred to as “mobile elements” because of their capacity to move around the genome. TEs are comprised of two larger categories, the transposons and retrotransposons. Transposons are segments of DNA which can excise themselves from a local sequence, move elsewhere, and splice themselves back in. Retrotransposons, on the other hand, don’t really move around but instead make RNA copies of themselves (usually when a nearby gene is also being transcribed); these RNA copies are then reverse transcribed into free-floating DNA which then, like the DNA transposon, splices itself back in (or piggybacks the splicing machinery of another retrotransposable element) [for review, see 1].

An extremely simplified representation of retrotransposition.
The human genome contains approximately 50% mobile element content. However, the vast majority of that content is extinct, meaning that is is no longer able to move around the genome. This happens due to the accumulation of mutations in the TE sequence which ultimately prevents it either from excising/copying itself or reinserting. In addition, the majority of TEs reside in intronic (i.e., non-coding) regions of DNA.
In our study, we used a bioinformatics approach to investigate TE content in high-risk autism-related genes. A gene list was derived from the database, AutismKB, and then compared to a listing of randomized control genes (for full content, see the end of our publication). What we found was that not only were autism-related genes more likely to house TE content, but they tended to house large numbers of TEs (100+).
Just to be clear, the majority of these TEs which were the subject of our study are extinct, meaning they’re not technically “mobile” anymore. So when discussing the relevance of this study to autism, we’re not talking about the majority of related mutations occurring because segments of genome are jumping around and promoting gene instability during insertion. What we are talking about though is how the extinct TE content, the characteristics of the sequences themselves, may be affecting rates of mutation in these genes.
In order to understand this, one really has to picture a gene as a 3D object whose tendency towards stability is defined by its shape. It’s shape is in turn determined by its DNA sequence (of which these TEs are an important part) and its interaction with molecular partners that regulate it, like proteins, RNA, and even metal ions like zinc. Since rates of mutation are driven by all these factors combined, you can imagine that the presence or absence of TE sequences in a given gene would definitely alter its overall shape and thereby affect the rates of mutation for that gene as well as the partners that are able to bind to it.
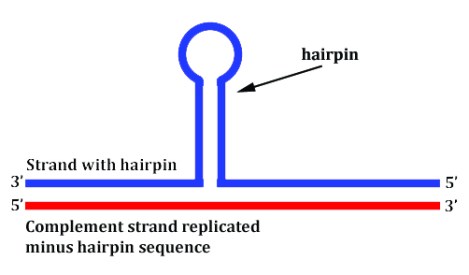
The funny thing about transposable elements is that they themselves tend to form unstable configurations. And when inserted into a genome, if the local DNA comes under any torsional strain such as occurs during transcription (gene expression), then these little TE sequences are known for jutting out of the double helix and forming outcroppings, like hairpin structures, triple helices, cruciforms, and other random coils. Look at the image below used as an analogy. In the lower portion of the image, the rubberband is placed under greater strain compared to the rubberband above it.
In the case of TE sequences in DNA, they have more of a tendency to break with the double helix under strain and form single strand (and some double-stranded) configurations which jut out of the traditional conformation, e.g., this next image.
Four problems can occur when this happens:
- When the DNA is being replicated either into DNA or RNA, you don’t necessarily get a reliable copy of the original because the replication machinery can actually skip structures like hairpins as if they weren’t even there, like in the image above.
- Alternatively, if the hairpin forms in just the wrong place, the replication machinery can actually stall and gene expression fails.
- When structures like hairpins or random coils form in the DNA during replication, this can also potentially lead to inappropriate expansion of certain segments, especially repetitive sequences (think of the CGG repeat expansions associated with Fragile X Syndrome).
- When alternative structures like hairpins form, there is some evidence to suggest that they may get “stuck” and need to be cut out of the DNA and repaired, potentially leading to inaccurate repair of the original sequence [2, 3].
The interesting thing about segments of DNA which house numerous TEs is that they seem to have attracted even more TEs over evolutionary time. If you look at any given gene with intronic TE content, usually you will find all types of mobile elements contained therein, which is surprising considering the different sequences these elements individually target for insertion. Not only does it suggest that coding regions may have a hard time accommodating TE insertion ultimately selecting against it, but it also suggests that introns attract TE insertion. And the more TEs, the more TEs they attract over the epochs. And the greater the TE content, the more unstable that region is overall. Just look at some of the largest genes in the human genome known for high rates of mutation, several of them also high-risk autism genes like CNTNAP2 and DMD, which house thousands of intronic mobile elements.
Scientists have traditionally been most interested in the coding regions of DNA, waving away intronic happenstances with the assumption that introns just don’t matter that much. But our study suggests differently. It suggests that the entire gene is important when it comes to stability and that introns can be an indicator of that gene’s history as well as its modern-day tendency to mutate.
So ultimately, I guess the question becomes: How does this knowledge help us in understanding autism? This kind of knowledge can aid us not only in dividing autisms into their many complex causations, genetic, epigenetic, and ecological alike, but also perhaps in seeing commonalities amongst them. And so while it’s currently difficult to predict what kinds of interventions may eventually be made available through such knowledge, it’s certain that the more we do know the greater means we’ll have to offer for improving the lives of autistic people.
And, yes, that’s a really fancy shmancy way of me saying “I don’t really know”. But we need to figure out as much as we can so that we can have as many options at our disposal as possible. You never quite know where science is going to take you until you go there.
NOTE: For anyone who would like to read the original publication and can’t get access, please email me and I’d be happy to send you a copy.



Congratulations on the publication.
Thanks very much, jaksichja. I hope ultimately the work proves useful. Will keep fingers crossed. 😉
Thanks for this post Emily (and for the work!). I haven’t had time to read your paper yet in great detail but am wondering if your findings might somehow link into increased HERV expression in autism http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0048831
any thoughts?
re voltage gated calcium channel genes, are you aware of this (unpublished) http://autismcalciumchannelopathy.com esp http://autismcalciumchannelopathy.com/Infectious_Agents.html
( I could speculate forever 😉 )
Fingers definitely crossed!
Natasa, I’ve been recently made aware of this study and am stilling mulling it over. At first glance, I suspect that higher expression levels on average in ASC of HERV-W and -H subgroups would probably indicate altered gene regulation, possibly through impaired mechanisms OR increased transcription of sites that house these viral sequences, since either could feasibly cause the results seen. So on some level it would definitely relate to aspects of our work, although moreso in those aspects of epigenetic regulation rather than the genetics we focused on here. Though I only alluded to it in the above, the act of transcription itself increases the likelihood of mutation in the gene being expressed because the transcription event increases local torsional strain. In a gene which may be “overexpressed”, you might expect to see higher rates of mutations in it.
Regarding calcium genes, in general some of these sequences are relatively unstable. Most of the CACNA series of genes house large TE content and they have a fairly extensive related family, suggesting they have been very prone to all types of mutations over time. Exactly how changes in calcium gene activity might lend towards an autism phenotype, I could only guess. Further gene network studies should help address those issues and some of them are staring to be done, so we can be hopeful of finding out in the coming years. 🙂
I second the congratulations. I feel proud of you, and grateful for a window on your talents, your work and your insights. When I need to remind myself of all that is vital and best about contemporary science and about working scientists, you and your site are what comes immediately to mind. Bravo for your work here. I’m printing this one, too.
:hooray:
Aw, thank you very much, proximity, for your EXCEPTIONALLY kind words. You likewise give me hope that the modern scientific mind is not completely devoid of romanticism and a dose of idealism. 😉
Pingback: Our Latest Study: The Link between Mobile Element Content and Autism-risk Genes | Cortical Chauvinism·
Thanks Emily! I’ve just come across this and find it fascinating, any thoughts if could be related to autism findings?
… The notion that somatic cells are free of new retrotransposition events is slowly changing. The development of better detection tools, such as synthetic L1 elements carrying reporter genes, is revealing the likelihood of retrotransposition in several somatic tissues (62,63). If the behavior of artificial L1s used in these studies is similar to that of endogenous elements, each individual can be considered to be a living library of de novo insertions, allowing stochastic gene expression in every single cell of the body. Of interest, we recently showed that L1 retrotransposition might have an insertional preference for neuronal genes when active in progenitor cells during the neuronal differentiation process (64). Newly generated L1 insertions can affect gene expression and influence neuronal fate. Such an observation indicates that neurons are a genetic mosaic for L1 content and that phenomenon may contribute to neuronal diversity (65). It remains to be determined the actual amount of endogenous mobilization, if such genetic mosaicism can influence neuronal networks and, as a result, have a functional consequence on behavior (66).
http://hmg.oxfordjournals.org/content/16/R2/R159.full#ref-64
http://www.ncbi.nlm.nih.gov/pubmed/15959507?dopt=Abstract
Hi, Natasa. Yes, that could definitely be relevant to our study in that autism-risk genes may, on average, be prime targets for L1 (or Alu) insertion events given their relative instability. However, of the mutation events which do occur, I suspect that retrotransposition would make up a minority. Meanwhile, mutations arising from general instability of these genes (CNVs, microsatellite expansion/contraction, and related SNPs) may be much more common occurrences. In addition, please keep in mind that only a small minority of autistic people actually house any of these potentially relevant mutations. For instance, studies on CNV occurrence have hovered around 6% of their samples. So it still seems that even though certain mutations may be more common in the population, most autistics do not appear to exhibit any relevant and identifiable genotype. Suggesting that a majority of these cases may be epigenetic/ecological in origin, even though they tend to be highly heritable. All of this of course doesn’t address the possibility of somatic mutations that may not be apparent in the blood, which is the tissue sample more studies select.
regarding “how changes in calcium gene activity might lend towards an autism phenotype” ‘Pediatric Neuroaids’ could be a good model to start with. Also some good references and ideas on how this might work on the link I provided …
Excellent and congratulations! One questiom, does this process continue throught one’s lifetime?
Yes, and has been better studied in the fields of cancer research. There’s a surprising amount of overlap, except in the case of cancer in which these types of issues tend to be cumulative over a lifetime, whereas in autism we’re talking about before, at, or just shortly after conception.
Emily, when you say that majority of autism cases ‘tend to be highly heritable’ I suppose what you referring to here is the higher concordance rates found in monozygotic twins … but isn’t this notion of ‘heritability of autism’ turning into a bit of a myth and in need of some serious re-branding and new terminology, considering that the high concordance rates, autism running in families etc are very likely due to shared genetic vulnerability to environmental insults. Take for example prenatal infection as etiological model for autism, where one’s genetic make-up will determine the risk and level of neurological damage and long term consequences … the findings by Saxena and colleagues are particularly interesting in this context don’t you think! http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0048835
Actually, I’m of the mind that the concept of “inheritance” needs to be better defined. Any form of inheritance cross-generationally could be considered “inherited” even if it’s something environmental. So part of inheritance is indeed consistency within the environment (and thank goodness because heaven forbid we fail to inherit something as necessary as sunlight or oxygen!). I think it requires breaking down different forms of inheritance into different modes. The genetic mode of inheritance for autism appears to be less stable compared to broader definitions of autism inheritance. Understanding the biology, how dynamic, interactive, and mutable DNA is, how the epigenome behaves and “remembers” and orchestrates, and how thin the line between self and environment at the level of the cell all but disappears… these are vital in redefining “inheritance” which is really an abstract concept, not biological or evolutionary. So, yes, a bit of a paradigm shift is warranted. The classic understanding of inheritance is exceedingly outdated in many aspects of biological science.
Natasa;
Twin studies in autism where one or both twins met diagnostic criteria for strictly defined or broadly defined autism have produced monozygotic (MZ) twin concordance rates as high as 92% in broadly defined autism. Classical twin study design heritability estimates cannot control for the high rates of de novo mutations in autism therefore twin study heritability estimates are significantly inflated.
I understood a smidgen of this article.� Maybe the first few layers of the onion.� However, I do know (as a trained child psychologist) the earlier we can intervene in early childhood with appropriate therapies the better off the child will be.� Good for you dear for the work you are doing. I can call you dear because I am
Mum
________________________________